Visualize the results of an emfx call.
Arguments
- x
An
emfxobject.- type
Character. The type of plot display. One of
"pointrange"(default),"errorbar", or"ribbon".- pch
Integer or character. Which plotting character or symbol to use (see
points). Defaults to 16 (i.e., small solid circle). Ignored iftype = "ribbon".- zero
Logical. Should 0-zero line be emphasized? Default is
TRUE.- grid
Logical. Should a background grid be displayed? Default is
TRUE.- ref
Integer. Reference line marker for event-study plot. Default is
-1(i.e., the period immediately preceding treatment). To remove completely, set toNA,NULL, orFALSE. Only used if the underlying object was computed usingemfx(..., type = "event").- ...
Additional arguments passed to
tinyplot::tinyplot.
Examples
# \dontrun{
# We’ll use the mpdta dataset from the did package (which you’ll need to
# install separately).
# install.packages("did")
data("mpdta", package = "did")
#
# Basic example
#
# The basic ETWFE workflow involves two consecutive function calls:
# 1) `etwfe` and 2) `emfx`
# 1) `etwfe`: Estimate a regression model with saturated interaction terms.
mod = etwfe(
fml = lemp ~ lpop, # outcome ~ controls (use 0 or 1 if none)
tvar = year, # time variable
gvar = first.treat, # group variable
data = mpdta, # dataset
vcov = ~countyreal # vcov adjustment (here: clustered by county)
)
# mod ## A fixest model object with fully saturated interaction effects.
# 2) `emfx`: Recover the treatment effects of interest.
(mod_es = emfx(mod, type = "event")) # dynamic ATE a la an event study
#>
#> event Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
#> 0 -0.0332 0.0134 -2.48 0.013 6.3 -0.0594 -0.00702
#> 1 -0.0573 0.0171 -3.34 <0.001 10.2 -0.0910 -0.02373
#> 2 -0.1379 0.0308 -4.48 <0.001 17.0 -0.1982 -0.07753
#> 3 -0.1095 0.0323 -3.39 <0.001 10.5 -0.1729 -0.04620
#>
#> Term: .Dtreat
#> Type: response
#> Comparison: TRUE - FALSE
#>
# Etc. Other aggregation type options are "simple" (the default), "group"
# and "calendar"
# To visualize results, use the native plot method (see `?plot.emfx`)
plot(mod_es)
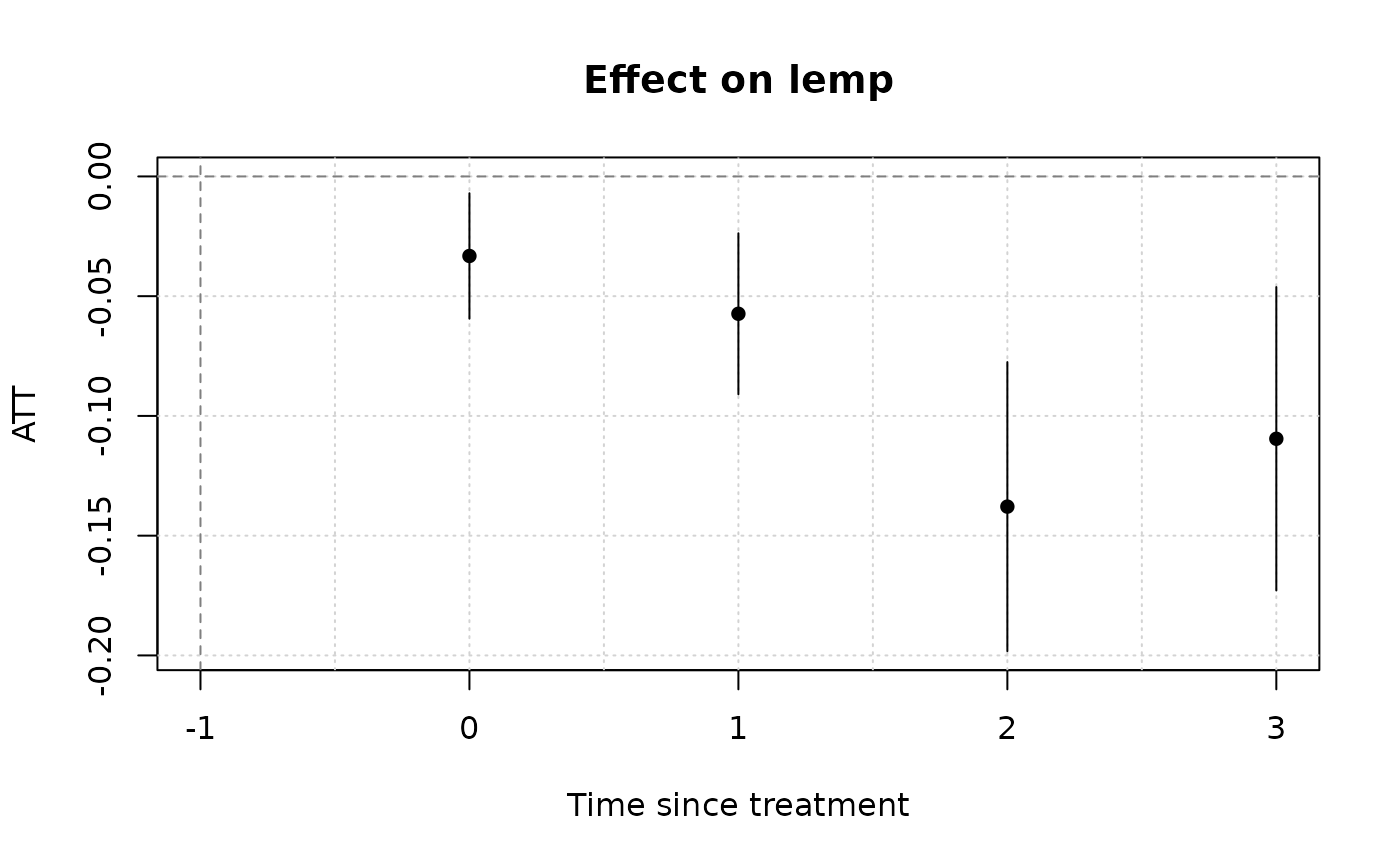 # Notice that we don't get any pre-treatment effects with the default
# "notyet" treated control group. Switch to the "never" treated control
# group if you want this.
etwfe(
lemp ~ lpop, tvar = year, gvar = first.treat, data = mpdta,
vcov = ~countyreal,
cgroup = "never" ## <= use never treated group as control
) |>
emfx("event") |>
plot()
# Notice that we don't get any pre-treatment effects with the default
# "notyet" treated control group. Switch to the "never" treated control
# group if you want this.
etwfe(
lemp ~ lpop, tvar = year, gvar = first.treat, data = mpdta,
vcov = ~countyreal,
cgroup = "never" ## <= use never treated group as control
) |>
emfx("event") |>
plot()
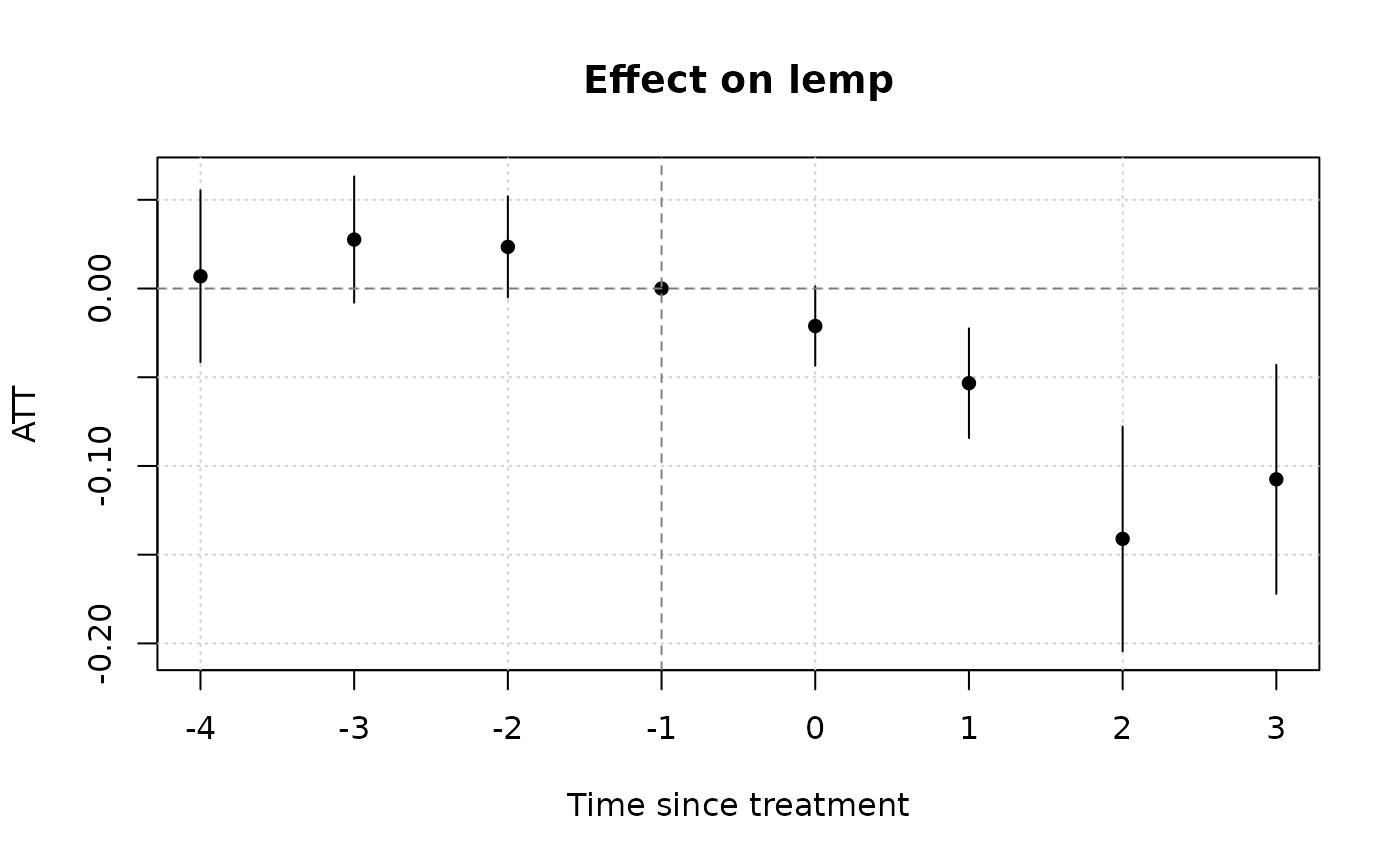 #
# Heterogeneous treatment effects
#
# Example where we estimate heterogeneous treatment effects for counties
# within the 8 US Great Lake states (versus all other counties).
gls = c("IL" = 17, "IN" = 18, "MI" = 26, "MN" = 27,
"NY" = 36, "OH" = 39, "PA" = 42, "WI" = 55)
mpdta$gls = substr(mpdta$countyreal, 1, 2) %in% gls
hmod = etwfe(
lemp ~ lpop, tvar = year, gvar = first.treat, data = mpdta,
vcov = ~countyreal,
xvar = gls ## <= het. TEs by gls
)
# Heterogeneous ATEs (could also specify "event", etc.)
emfx(hmod)
#>
#> .Dtreat gls Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
#> TRUE FALSE -0.0600 0.0344 -1.74 0.0811 3.6 -0.127 0.00741
#> TRUE TRUE -0.0449 0.0281 -1.60 0.1101 3.2 -0.100 0.01018
#>
#> Term: .Dtreat
#> Type: response
#> Comparison: TRUE - FALSE
#>
# To test whether the ATEs across these two groups (non-GLS vs GLS) are
# statistically different, simply pass an appropriate "hypothesis" argument.
emfx(hmod, hypothesis = "b1 = b2")
#>
#> Hypothesis Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
#> b1=b2 -0.0151 0.0538 -0.28 0.779 0.4 -0.121 0.0904
#>
#> Type: response
#>
plot(emfx(hmod))
#
# Heterogeneous treatment effects
#
# Example where we estimate heterogeneous treatment effects for counties
# within the 8 US Great Lake states (versus all other counties).
gls = c("IL" = 17, "IN" = 18, "MI" = 26, "MN" = 27,
"NY" = 36, "OH" = 39, "PA" = 42, "WI" = 55)
mpdta$gls = substr(mpdta$countyreal, 1, 2) %in% gls
hmod = etwfe(
lemp ~ lpop, tvar = year, gvar = first.treat, data = mpdta,
vcov = ~countyreal,
xvar = gls ## <= het. TEs by gls
)
# Heterogeneous ATEs (could also specify "event", etc.)
emfx(hmod)
#>
#> .Dtreat gls Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
#> TRUE FALSE -0.0600 0.0344 -1.74 0.0811 3.6 -0.127 0.00741
#> TRUE TRUE -0.0449 0.0281 -1.60 0.1101 3.2 -0.100 0.01018
#>
#> Term: .Dtreat
#> Type: response
#> Comparison: TRUE - FALSE
#>
# To test whether the ATEs across these two groups (non-GLS vs GLS) are
# statistically different, simply pass an appropriate "hypothesis" argument.
emfx(hmod, hypothesis = "b1 = b2")
#>
#> Hypothesis Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
#> b1=b2 -0.0151 0.0538 -0.28 0.779 0.4 -0.121 0.0904
#>
#> Type: response
#>
plot(emfx(hmod))
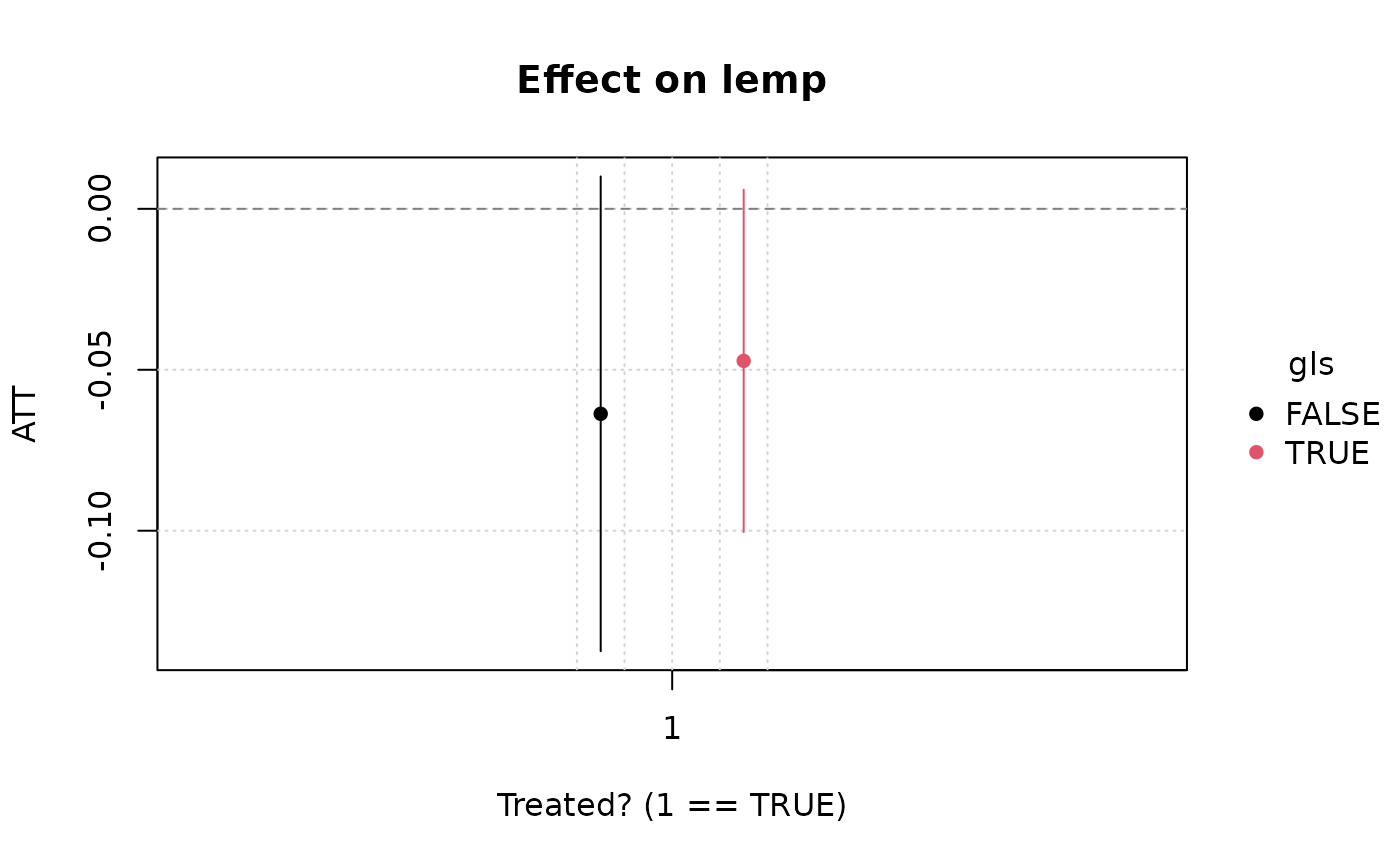 #
# Nonlinear model (distribution / link) families
#
# Poisson example
mpdta$emp = exp(mpdta$lemp)
etwfe(
emp ~ lpop, tvar = year, gvar = first.treat, data = mpdta,
vcov = ~countyreal,
family = "poisson" ## <= family arg for nonlinear options
) |>
emfx("event")
#> The variables '.Dtreat:first.treat::2006:year::2004',
#> '.Dtreat:first.treat::2006:year::2005', '.Dtreat:first.treat::2007:year::2004',
#> '.Dtreat:first.treat::2007:year::2005', '.Dtreat:first.treat::2007:year::2006',
#> '.Dtreat:first.treat::2006:year::2004:lpop_dm' and 4 others have been removed
#> because of collinearity (see $collin.var).
#>
#> event Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
#> 0 -25.35 15.9 -1.5957 0.11056 3.2 -56.5 5.79
#> 1 1.09 40.3 0.0271 0.97838 0.0 -77.9 80.07
#> 2 -75.12 23.2 -3.2445 0.00118 9.7 -120.5 -29.74
#> 3 -101.82 27.1 -3.7590 < 0.001 12.5 -154.9 -48.73
#>
#> Term: .Dtreat
#> Type: response
#> Comparison: TRUE - FALSE
#>
# }
#
# Nonlinear model (distribution / link) families
#
# Poisson example
mpdta$emp = exp(mpdta$lemp)
etwfe(
emp ~ lpop, tvar = year, gvar = first.treat, data = mpdta,
vcov = ~countyreal,
family = "poisson" ## <= family arg for nonlinear options
) |>
emfx("event")
#> The variables '.Dtreat:first.treat::2006:year::2004',
#> '.Dtreat:first.treat::2006:year::2005', '.Dtreat:first.treat::2007:year::2004',
#> '.Dtreat:first.treat::2007:year::2005', '.Dtreat:first.treat::2007:year::2006',
#> '.Dtreat:first.treat::2006:year::2004:lpop_dm' and 4 others have been removed
#> because of collinearity (see $collin.var).
#>
#> event Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
#> 0 -25.35 15.9 -1.5957 0.11056 3.2 -56.5 5.79
#> 1 1.09 40.3 0.0271 0.97838 0.0 -77.9 80.07
#> 2 -75.12 23.2 -3.2445 0.00118 9.7 -120.5 -29.74
#> 3 -101.82 27.1 -3.7590 < 0.001 12.5 -154.9 -48.73
#>
#> Term: .Dtreat
#> Type: response
#> Comparison: TRUE - FALSE
#>
# }