Companion function to etwfe, enabling the recovery of aggregate treatment
effects along different dimensions of interest (e.g, an event study of
dynamic average treatment effects). emfx is a light wrapper around the
slopes function from the marginaleffects
package.
Arguments
- object
An
etwfemodel object.- type
Character. The desired type of post-estimation aggregation.
- by_xvar
Logical. Should the results account for heterogeneous treatment effects? Only relevant if the preceding
etwfecall included a specifiedxvarargument, i.e. interacted categorical covariate. The default behaviour ("auto") is to automatically estimate heterogeneous treatment effects for each level ofxvarif these are detected as part of the underlyingetwfemodel object. Users can override by setting to eitherFALSEorTRUE.See the "Heterogeneous treatment effects" section below.- compress
Logical. Compress the data by (period by cohort) groups before calculating marginal effects? This trades off a slight loss in precision (typically around the 1st or 2nd significant decimal point) for a substantial improvement in estimation time for large datasets. The default behaviour (
"auto") is to automatically compress if the original dataset has more than 500,000 rows. Users can override by setting eitherFALSEorTRUE. Note that collapsing by group is only valid if the precedingetwfecall was run with"ivar = NULL"(the default). See the "Performance tips" section below.- collapse
Logical. An alias for
compress(only used for backwards compatability and ignored if both arguments are provided). The behaviour is identical, but it will trigger a message nudging users to rather use thecompressargument.- predict
Character. The type (scale) of prediction used to compute the marginal effects. If
"response"(the default), then the output is at the level of the response variable, i.e. it is the expected predictor \(E(Y|X)\). If"link", the value returned is the linear predictor of the fitted model, i.e. \(X\cdot \beta\). The difference should only matter for nonlinear models. (Note: This argument is typically calledtypewhen use inpredictorslopes, but we rename it here to avoid a clash with the top-leveltypeargument above.)- post_only
Logical. Drop pre-treatment ATTs? Only evaluated if (a)
type = "event"and (b) the originaletwfemodel object was estimated using the default"notyet"treated control group. If conditions (a) and (b) are met then the pre-treatment effects will be zero as a mechanical result of ETWFE's estimation setup. The default behaviour (TRUE) is thus to drop these nuisance rows from the dataset. Thepost_onlyargument recognises that you may still want to keep them for presentation purposes (e.g., plotting an event study). Nevertheless, be forewarned that enabling that behaviour viaFALSEis strictly performative: the "zero" treatment effects for any pre-treatment periods is purely an artefact of the estimation setup.- window
Numeric of length 1 or 2. Limits the temporal window of consideration around treatment.
NULL (default): Include all available periods.
Length 1: Truncate to a symmetric window around the treatment event. E.g.,
window = 2will truncate to two pre-treatment periods and two post-treatment periods.Length 2: Asymmetric window, where the first number gives the maximum number of pre-treatment periods and the second number gives the maximum number of post-treatment periods. E.g.,
window = c(5, 2)will truncate to five pre-treatment periods and two post-treatment periods.
Note that the pre-treatment truncation is only ever binding in cases where the "never" treated group is used as a control, i.e.,
etwfe(..., cgroup = "never")in the original call.- lean
Logical. Default is
FALSE. Switching toTRUEenforces a lean return object; namely a simple data.frame of the main results, stripped of ancillary attributes. Note that this will disable some advancedmarginaleffectspost-processing features, but those are unlikely to be used in theemfxcontext. The upside is a potentially dramatic reduction in the size of the return object. Consequently, we may change the default toTRUEin a future version of etwfe.- ...
Additional arguments passed to
marginaleffects::slopes. For example, you can passvcov = FALSEto dramatically speed up estimation times of the main marginal effects (but at the cost of not getting any information about standard errors; see Performance tips below). Another potentially useful application is testing whether heterogeneous treatment effects (i.e., the levels of anyxvarcovariate) are equal by invoking thehypothesisargument, e.g.hypothesis = "b1 = b2".
Value
A data.frame of aggregated treatment effects along the
dimension(s) of interested. Note that this data.frame will have been
overloaded with the slopes class, and so
will come with a special print method. But the underlying columns will
usually include:
termcontrast<type>(i.e., the name of yourtypestring)estimatestd.errorstatisticp.values.valueconf.lowconf.high
Performance tips
Under most situations, etwfe should complete very quickly. For its part,
emfx is quite performant too and should take a few seconds or less for
datasets under 100k rows. However, emfx's computation time does tend to
scale non-linearly with the size of the original data, as well as the
number of interactions from the underlying etwfe model. Without getting
too deep into the weeds, the numerical delta method used to recover the
ATEs of interest has to estimate two prediction models for each
coefficient in the model and then compute their standard errors. So, it's
a potentially expensive operation that can push the computation time for
large datasets (> 1m rows) up to several minutes or longer.
Fortunately, there are two complementary strategies that you can use to
speed things up. The first is to turn off the most expensive part of the
whole procedure—standard error calculation—by calling emfx(..., vcov = FALSE). Doing so should bring the estimation time back down to a few
seconds or less, even for datasets in excess of a million rows. While the
loss of standard errors might not be an acceptable trade-off for projects
where statistical inference is critical, the good news is this first
strategy can still be combined our second strategy. It turns out that
collapsing the data by groups prior to estimating the marginal effects can
yield substantial speed gains of its own. Users can do this by invoking
the emfx(..., collapse = TRUE) argument. While the effect here is not as
dramatic as the first strategy, our second strategy does have the virtue
of retaining information about the standard errors. The trade-off this
time, however, is that collapsing our data does lead to a loss in accuracy
for our estimated parameters. On the other hand, testing suggests that
this loss in accuracy tends to be relatively minor, with results
equivalent up to the 1st or 2nd significant decimal place (or even
better).
Summarizing, here's a quick plan of attack for you to try if you are worried about the estimation time for large datasets and models:
Estimate
mod = etwfe(...)as per usual.Run
emfx(mod, vcov = FALSE, ...).Run
emfx(mod, vcov = FALSE, collapse = TRUE, ...).Compare the point estimates from steps 1 and 2. If they are are similar enough to your satisfaction, get the approximate standard errors by running
emfx(mod, collapse = TRUE, ...).
Heterogeneous treatment effects
Specifying etwfe(..., xvar = <xvar>) will generate interaction effects
for all levels of <xvar> as part of the main regression model. The
reason that this is useful (as opposed to a regular, non-interacted
covariate in the formula RHS) is that it allows us to estimate
heterogeneous treatment effects as part of the larger ETWFE framework.
Specifically, we can recover heterogeneous treatment effects for each
level of <xvar> by passing the resulting etwfe model object on to
emfx().
For example, imagine that we have a categorical variable called "age" in
our dataset, with two distinct levels "adult" and "child". Running
emfx(etwfe(..., xvar = age)) will tell us how the efficacy of treatment
varies across adults and children. We can then also leverage the in-built
hypothesis testing infrastructure of marginaleffects to test whether
the treatment effect is statistically different across these two age
groups; see Examples below. Note the same principles carry over to
categorical variables with multiple levels, or even continuous variables
(although continuous variables are not as well supported yet).
References
Wong, Jeffrey et al. (2021). You Only Compress Once: Optimal Data Compression for Estimating Linear Models. Working paper (version: March 16, 2021). Available: https://doi.org/10.48550/arXiv.2102.11297
See also
marginaleffects::slopes which does the heavily lifting behind the
scenes. etwfe is the companion estimating function that should be run
before emfx.
Examples
# \dontrun{
# We’ll use the mpdta dataset from the did package (which you’ll need to
# install separately).
# install.packages("did")
data("mpdta", package = "did")
#
# Basic example
#
# The basic ETWFE workflow involves two consecutive function calls:
# 1) `etwfe` and 2) `emfx`
# 1) `etwfe`: Estimate a regression model with saturated interaction terms.
mod = etwfe(
fml = lemp ~ lpop, # outcome ~ controls (use 0 or 1 if none)
tvar = year, # time variable
gvar = first.treat, # group variable
data = mpdta, # dataset
vcov = ~countyreal # vcov adjustment (here: clustered by county)
)
# mod ## A fixest model object with fully saturated interaction effects.
# 2) `emfx`: Recover the treatment effects of interest.
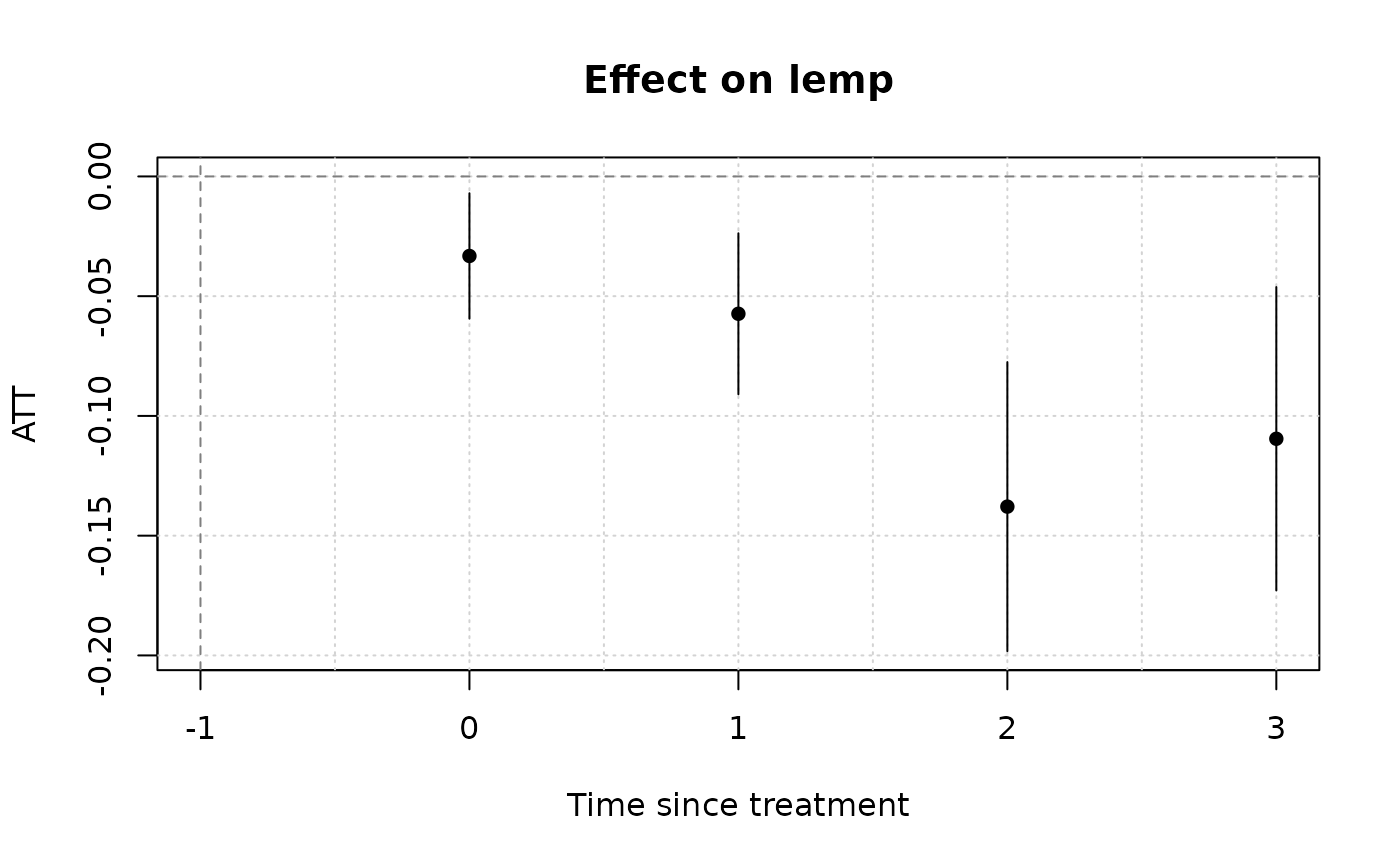
(mod_es = emfx(mod, type = "event")) # dynamic ATE a la an event study
#>
#> event Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
#> 0 -0.0332 0.0134 -2.48 0.013 6.3 -0.0594 -0.00702
#> 1 -0.0573 0.0171 -3.34 <0.001 10.2 -0.0910 -0.02373
#> 2 -0.1379 0.0308 -4.48 <0.001 17.0 -0.1982 -0.07753
#> 3 -0.1095 0.0323 -3.39 <0.001 10.5 -0.1729 -0.04620
#>
#> Term: .Dtreat
#> Type: response
#> Comparison: TRUE - FALSE
#>
# Etc. Other aggregation type options are "simple" (the default), "group"
# and "calendar"
# To visualize results, use the native plot method (see `?plot.emfx`)
plot(mod_es)
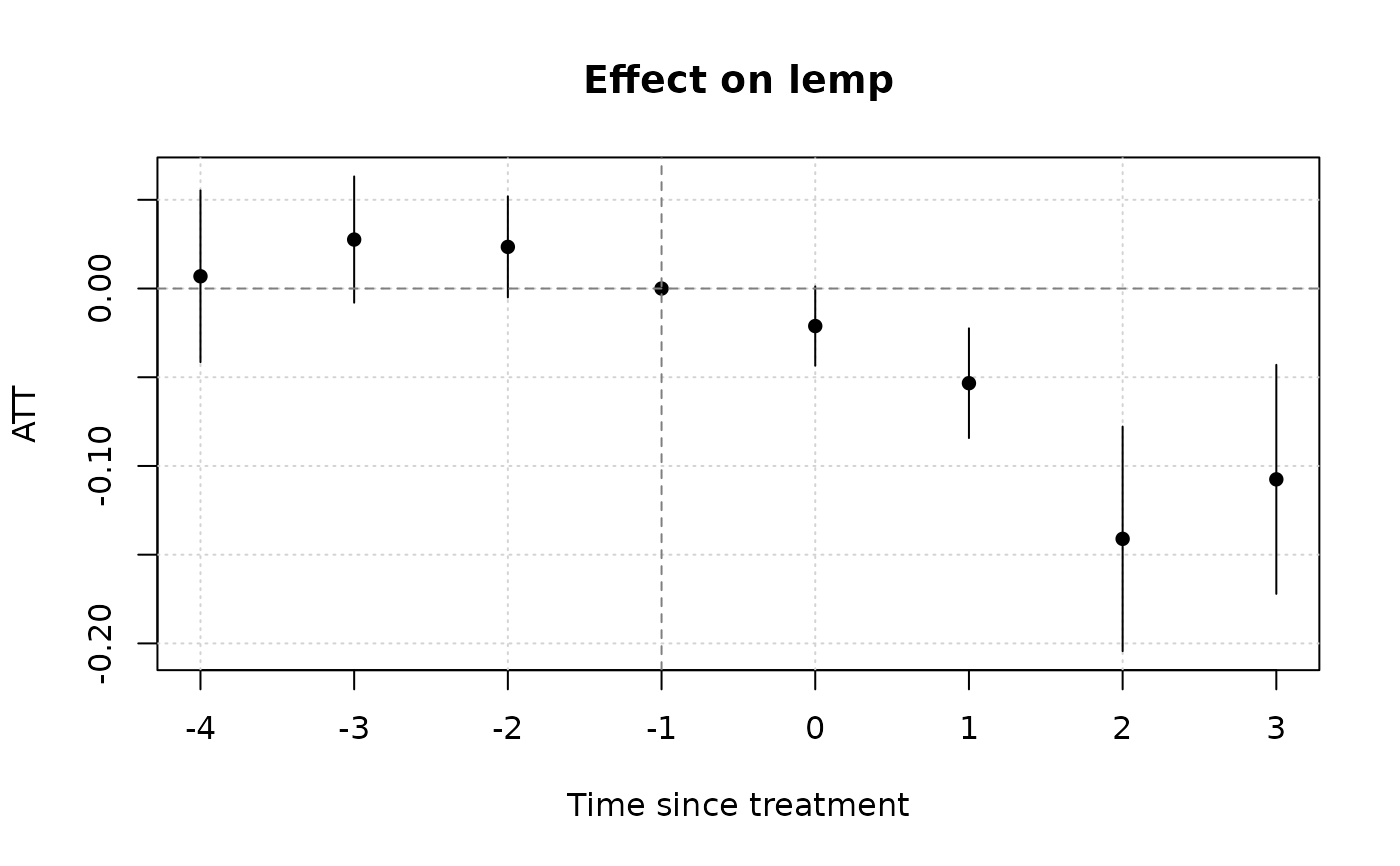
 # Notice that we don't get any pre-treatment effects with the default
# "notyet" treated control group. Switch to the "never" treated control
# group if you want this.
etwfe(
lemp ~ lpop, tvar = year, gvar = first.treat, data = mpdta,
vcov = ~countyreal,
cgroup = "never" ## <= use never treated group as control
) |>
emfx("event") |>
plot()
# Notice that we don't get any pre-treatment effects with the default
# "notyet" treated control group. Switch to the "never" treated control
# group if you want this.
etwfe(
lemp ~ lpop, tvar = year, gvar = first.treat, data = mpdta,
vcov = ~countyreal,
cgroup = "never" ## <= use never treated group as control
) |>
emfx("event") |>
plot()
 #
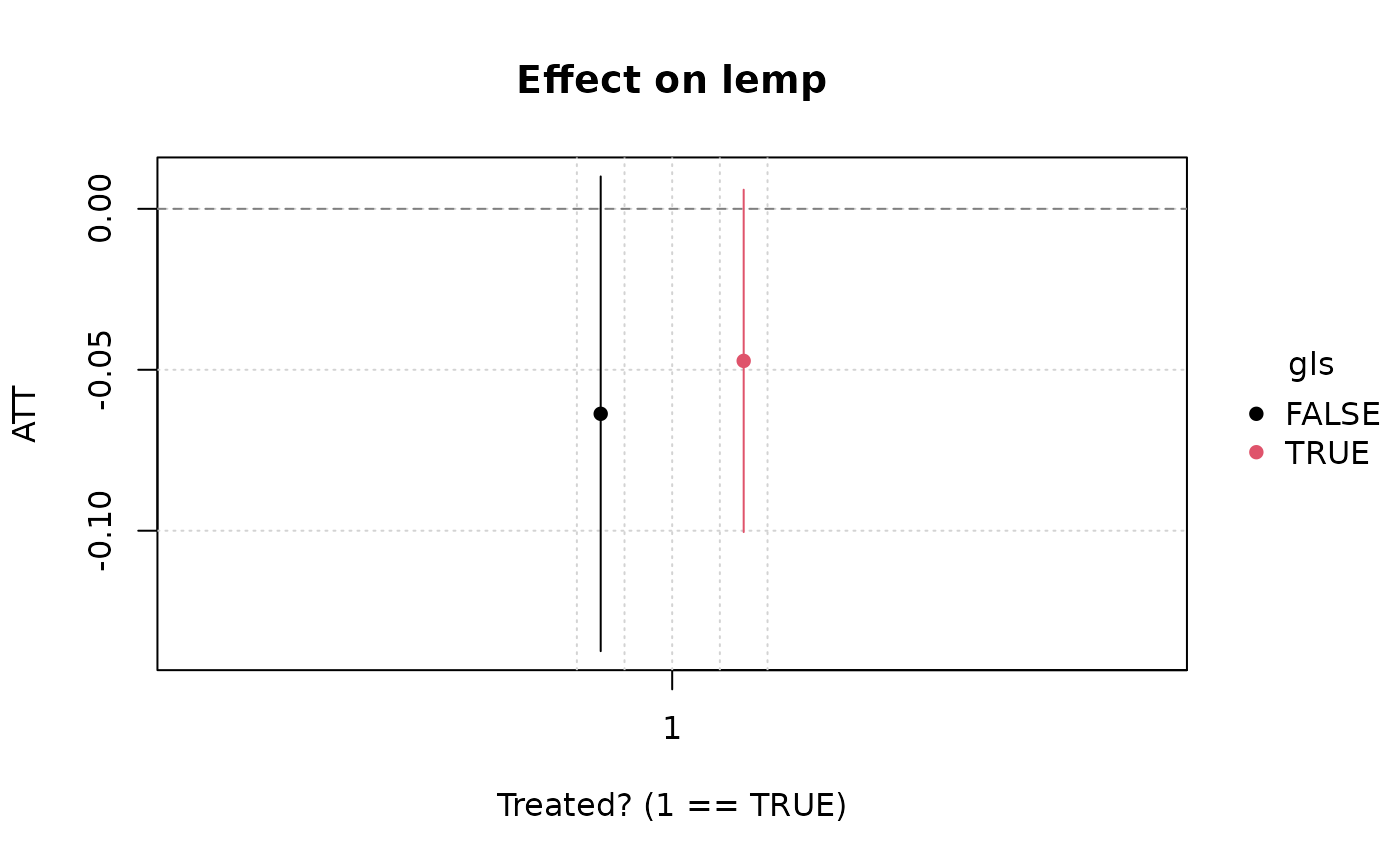
# Heterogeneous treatment effects
#
# Example where we estimate heterogeneous treatment effects for counties
# within the 8 US Great Lake states (versus all other counties).
gls = c("IL" = 17, "IN" = 18, "MI" = 26, "MN" = 27,
"NY" = 36, "OH" = 39, "PA" = 42, "WI" = 55)
mpdta$gls = substr(mpdta$countyreal, 1, 2) %in% gls
hmod = etwfe(
lemp ~ lpop, tvar = year, gvar = first.treat, data = mpdta,
vcov = ~countyreal,
xvar = gls ## <= het. TEs by gls
)
# Heterogeneous ATEs (could also specify "event", etc.)
emfx(hmod)
#>
#> .Dtreat gls Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
#> TRUE FALSE -0.0600 0.0344 -1.74 0.0811 3.6 -0.127 0.00741
#> TRUE TRUE -0.0449 0.0281 -1.60 0.1101 3.2 -0.100 0.01018
#>
#> Term: .Dtreat
#> Type: response
#> Comparison: TRUE - FALSE
#>
# To test whether the ATEs across these two groups (non-GLS vs GLS) are
# statistically different, simply pass an appropriate "hypothesis" argument.
emfx(hmod, hypothesis = "b1 = b2")
#> Warning:
#> It is essential to check the order of estimates when specifying hypothesis tests using positional indices like b1, b2, etc. The indices of estimates can change depending on the order of rows in the original dataset, user-supplied arguments, model-fitting package, and version of `marginaleffects`.
#>
#> It is also good practice to use assertions that ensure the order of estimates is consistent across different runs of the same code. Example:
#>
#> ```r
#> mod <- lm(mpg ~ am * carb, data = mtcars)
#>
#> # assertion for safety
#> p <- avg_predictions(mod, by = 'carb')
#> stopifnot(p$carb[1] == 1, p$carb[2] == 2)
#>
#> # hypothesis test
#> avg_predictions(mod, by = 'carb', hypothesis = 'b1 - b2 = 0')
#> ```
#>
#> Disable this warning with: `options(marginaleffects_safe = FALSE)`
#> This warning appears once per session.
#>
#> Hypothesis Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
#> b1=b2 -0.0151 0.0538 -0.28 0.779 0.4 -0.121 0.0904
#>
#> Type: response
#>
plot(emfx(hmod))
#
# Heterogeneous treatment effects
#
# Example where we estimate heterogeneous treatment effects for counties
# within the 8 US Great Lake states (versus all other counties).
gls = c("IL" = 17, "IN" = 18, "MI" = 26, "MN" = 27,
"NY" = 36, "OH" = 39, "PA" = 42, "WI" = 55)
mpdta$gls = substr(mpdta$countyreal, 1, 2) %in% gls
hmod = etwfe(
lemp ~ lpop, tvar = year, gvar = first.treat, data = mpdta,
vcov = ~countyreal,
xvar = gls ## <= het. TEs by gls
)
# Heterogeneous ATEs (could also specify "event", etc.)
emfx(hmod)
#>
#> .Dtreat gls Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
#> TRUE FALSE -0.0600 0.0344 -1.74 0.0811 3.6 -0.127 0.00741
#> TRUE TRUE -0.0449 0.0281 -1.60 0.1101 3.2 -0.100 0.01018
#>
#> Term: .Dtreat
#> Type: response
#> Comparison: TRUE - FALSE
#>
# To test whether the ATEs across these two groups (non-GLS vs GLS) are
# statistically different, simply pass an appropriate "hypothesis" argument.
emfx(hmod, hypothesis = "b1 = b2")
#> Warning:
#> It is essential to check the order of estimates when specifying hypothesis tests using positional indices like b1, b2, etc. The indices of estimates can change depending on the order of rows in the original dataset, user-supplied arguments, model-fitting package, and version of `marginaleffects`.
#>
#> It is also good practice to use assertions that ensure the order of estimates is consistent across different runs of the same code. Example:
#>
#> ```r
#> mod <- lm(mpg ~ am * carb, data = mtcars)
#>
#> # assertion for safety
#> p <- avg_predictions(mod, by = 'carb')
#> stopifnot(p$carb[1] == 1, p$carb[2] == 2)
#>
#> # hypothesis test
#> avg_predictions(mod, by = 'carb', hypothesis = 'b1 - b2 = 0')
#> ```
#>
#> Disable this warning with: `options(marginaleffects_safe = FALSE)`
#> This warning appears once per session.
#>
#> Hypothesis Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
#> b1=b2 -0.0151 0.0538 -0.28 0.779 0.4 -0.121 0.0904
#>
#> Type: response
#>
plot(emfx(hmod))
 #
# Nonlinear model (distribution / link) families
#
# Poisson example
mpdta$emp = exp(mpdta$lemp)
etwfe(
emp ~ lpop, tvar = year, gvar = first.treat, data = mpdta,
vcov = ~countyreal,
family = "poisson" ## <= family arg for nonlinear options
) |>
emfx("event")
#> The variables '.Dtreat:first.treat::2006:year::2004',
#> '.Dtreat:first.treat::2006:year::2005', '.Dtreat:first.treat::2007:year::2004',
#> '.Dtreat:first.treat::2007:year::2005', '.Dtreat:first.treat::2007:year::2006',
#> '.Dtreat:first.treat::2006:year::2004:lpop_dm' and 4 others have been removed
#> because of collinearity (see $collin.var).
#>
#> event Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
#> 0 -25.35 15.9 -1.5957 0.11056 3.2 -56.5 5.79
#> 1 1.09 40.3 0.0271 0.97838 0.0 -77.9 80.07
#> 2 -75.12 23.2 -3.2445 0.00118 9.7 -120.5 -29.74
#> 3 -101.82 27.1 -3.7590 < 0.001 12.5 -154.9 -48.73
#>
#> Term: .Dtreat
#> Type: response
#> Comparison: TRUE - FALSE
#>
# }
#
# Nonlinear model (distribution / link) families
#
# Poisson example
mpdta$emp = exp(mpdta$lemp)
etwfe(
emp ~ lpop, tvar = year, gvar = first.treat, data = mpdta,
vcov = ~countyreal,
family = "poisson" ## <= family arg for nonlinear options
) |>
emfx("event")
#> The variables '.Dtreat:first.treat::2006:year::2004',
#> '.Dtreat:first.treat::2006:year::2005', '.Dtreat:first.treat::2007:year::2004',
#> '.Dtreat:first.treat::2007:year::2005', '.Dtreat:first.treat::2007:year::2006',
#> '.Dtreat:first.treat::2006:year::2004:lpop_dm' and 4 others have been removed
#> because of collinearity (see $collin.var).
#>
#> event Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
#> 0 -25.35 15.9 -1.5957 0.11056 3.2 -56.5 5.79
#> 1 1.09 40.3 0.0271 0.97838 0.0 -77.9 80.07
#> 2 -75.12 23.2 -3.2445 0.00118 9.7 -120.5 -29.74
#> 3 -101.82 27.1 -3.7590 < 0.001 12.5 -154.9 -48.73
#>
#> Term: .Dtreat
#> Type: response
#> Comparison: TRUE - FALSE
#>
# }